In this note, we show details and codes to reproduce results shown in the paper.
Deep learning enables accurate clustering with batch effect removal in single-cell RNA-seq analysis
Xiangjie Li1,2,3, Kui Wang1,4, Yafei Lyu1, Huize Pan5, Jingxiao Zhang2, Dwight Stambolian6, Katalin Susztak7, Muredach P. Reilly5, Gang Hu1,8*, Mingyao Li1*
Show more about authors
- Department of Biostatistics, Epidemiology and Informatics, University of Pennsylvania Perelman School of Medicine, Philadelphia, PA 19104, USA.
- Center for Applied Statistics, School of Statistics, Renmin University, Beijing, China.
- State Key Laboratory of Cardiovascular Disease, Fuwai Hospital, National Center for Cardiovascular Diseases, Chinese Academy of Medical Sciences and Peking Union Medical College, Beijing 100037, China.
- Department of Information Theory and Data Science, School of Mathematical Sciences and LPMC, Nankai University, Tianjin 300071, China.
- Cardiology Division, Department of Medicine, Columbia University Medical Center, New York, NY 10032, USA.
- Department of Ophthalmology, University of Pennsylvania Perelman School of Medicine, Philadelphia, PA 19104, USA.
- Departments of Medicine and Genetics, University of Pennsylvania Perelman School of Medicine, Philadelphia, PA 19104, USA.
- School of Statistics and Data Science, Key Laboratory for medical Data Analysis and Statistical Research of Tianjin, Nankai University, Tianjin 300071, China.
Figures
- Figures related to macaque retina data.
- Figures related to human pancreas data.
- Figures related to human PBMC data.
- Figures related to mouse bone marrow myeloid progenitor cell dataset.
- Figures related to human monocyte data.
- Figures related to running time and memory usage
- Figures related to 1.3 million brain cells from E18 mice.
The two figures in Figure 10 can be found in Figures related to running time and memory usage and Figures related to macaque retina data, respectively.
Workflow figure
Our workflow figure 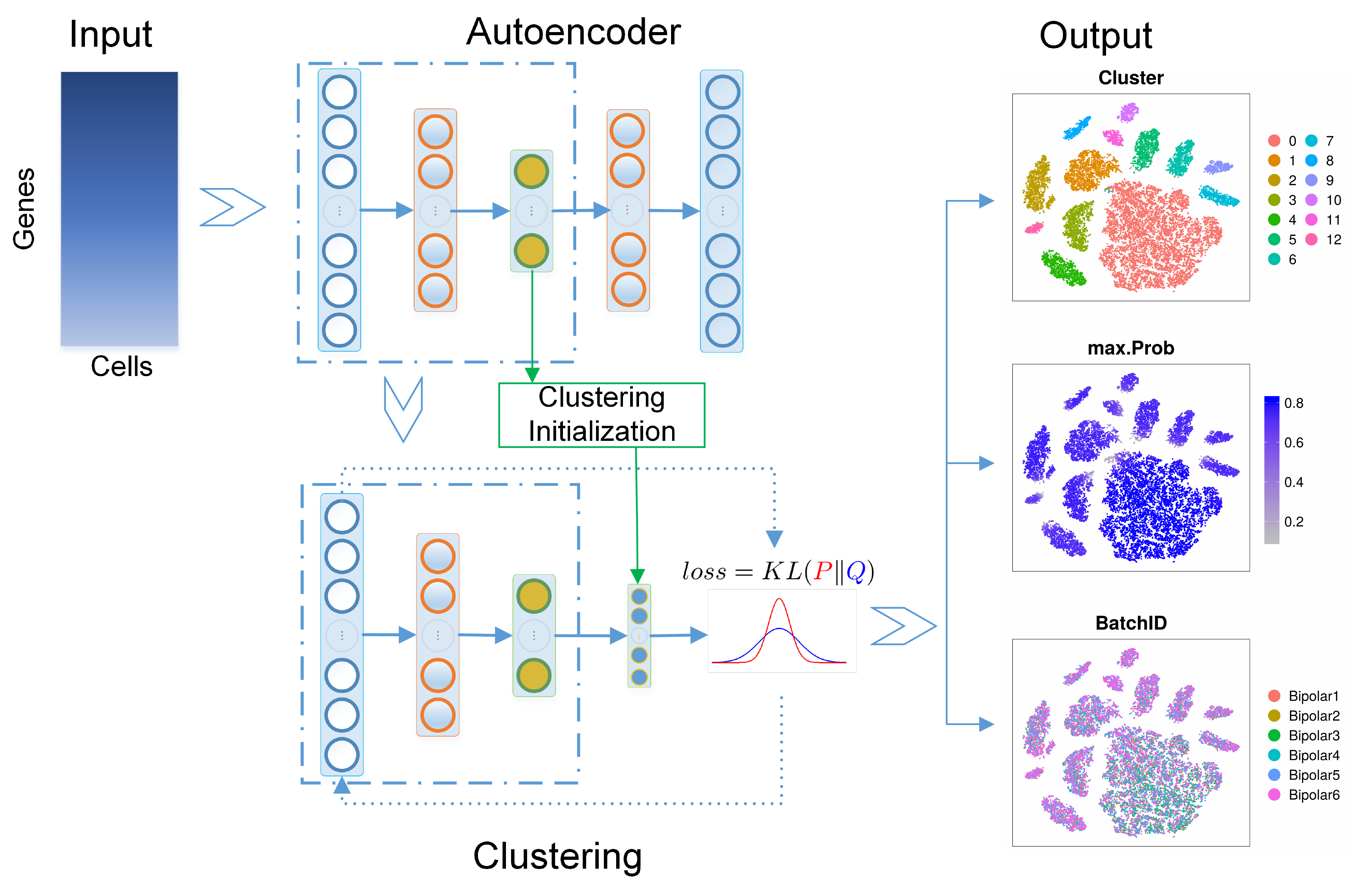 was designed by Microsoft Visio.
was designed by Microsoft Visio.
Please contact eleozzr@gmail.com or ele717@163.com with any questions about the desc package.